Examples
We have provided a concise description of our web server's functionality to enhance user convenience. The step-by-step guide illustrates the input process and the corresponding output of our web server.
Input description:
Our webserver takes single RNA sequence details as input at a time. You can write or copy paste the name of the RNA along with its sequence or can upload a file in Fasta format. Details of three different RNAs are given as Examples for your understanding. For uploading file, it should be in your local device.
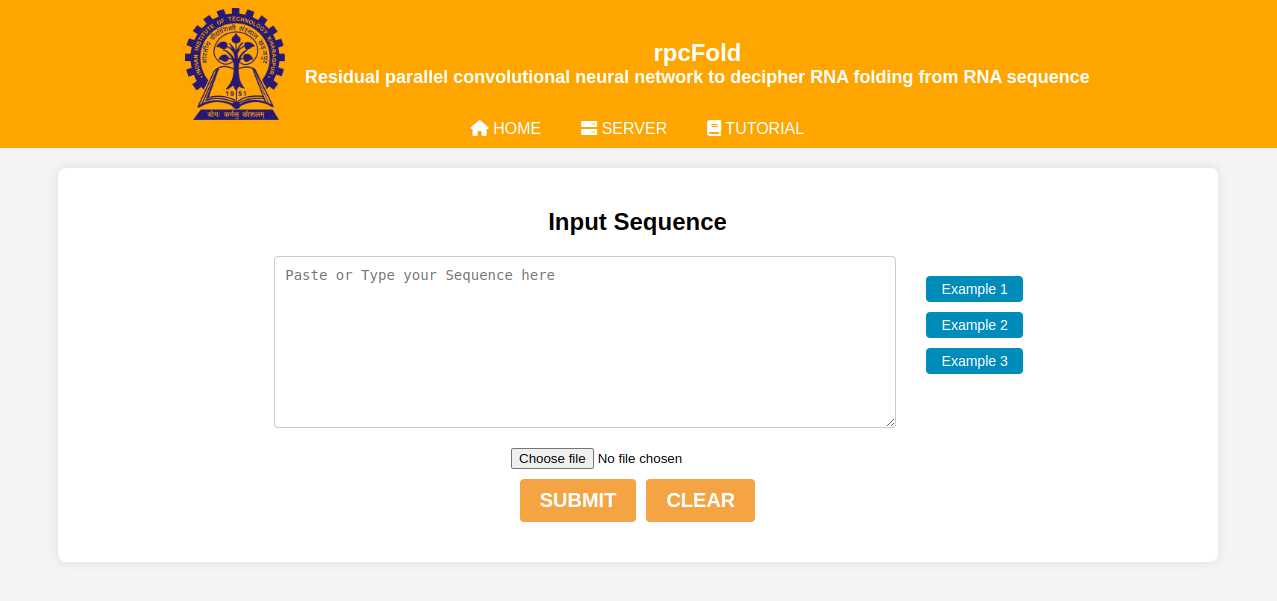
Figure 1: Input description example interface.
Input file processing:
We have added three examples for the better understanding of the input format. By clicking on any of the example you can see the name and corresponding sequence of the RNA. Now if you submit the RNA details, our server will start predicting the RNA secondary structure based on our pre-trained rpcFold model.You can also submit your input by uploading an RNA fasta file.The file should be available in your local device.After submitting your input, please wait. Based on the workload on our server it may take some time. Please don't reload the page while processing your file.
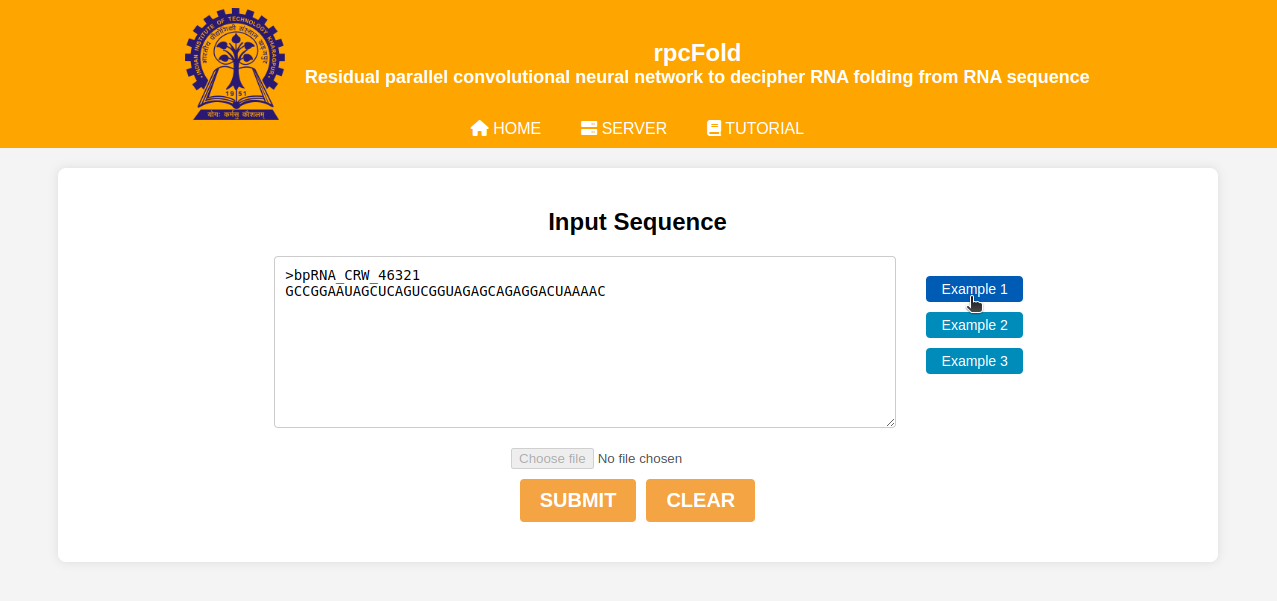
Figure 2: Input file processing example interface.
Output description:
The output of our server is the predicted RNA secondary structure from the given input sequence. Our output page contains the name of given input RNA, original input sequence and the predicted output structure in 'dot-bracket' notation. We have also incorporated the graphical visualization of the predicted structure using the Forna secondary structure visualization tool. You can also download the output result by clicking on the Download button on the right side of the page. The download file is in .txt format and contains the input name, sequence and predicted structure.
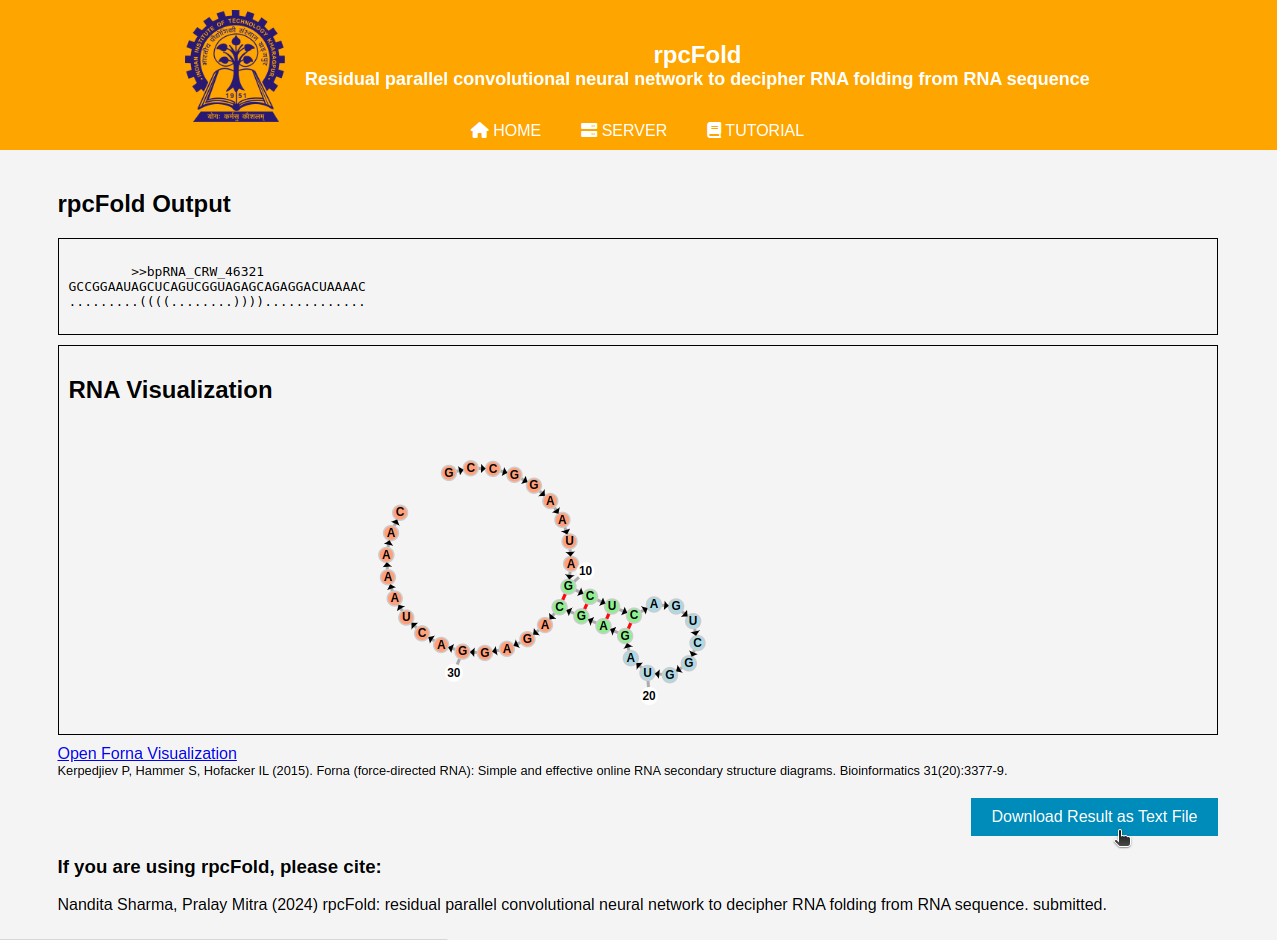
Figure 3: Output description example interface.



